
Musite, a Tool for Global Prediction of General and Kinase-specific Phosphorylation Sites - ScienceDirect

ProteoConnections: A bioinformatics platform to facilitate proteome and phosphoproteome analyses - Courcelles - 2011 - PROTEOMICS - Wiley Online Library
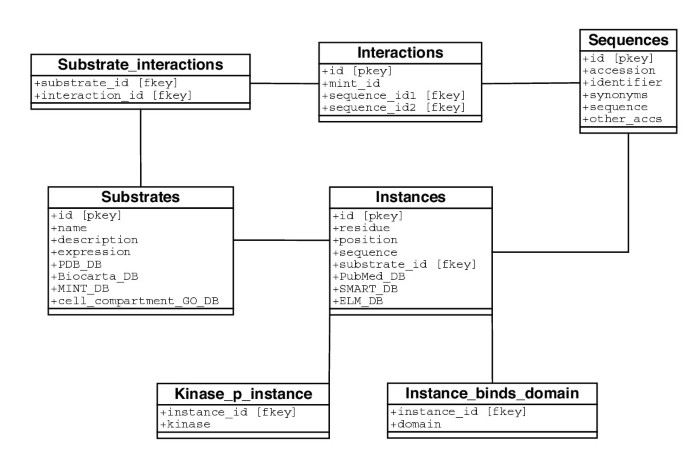
Phospho.ELM: A database of experimentally verified phosphorylation sites in eukaryotic proteins | BMC Bioinformatics | Full Text

Profiling the Human Phosphoproteome to Estimate the True Extent of Protein Phosphorylation | Journal of Proteome Research

IJMS | Free Full-Text | GasPhos: Protein Phosphorylation Site Prediction Using a New Feature Selection Approach with a GA-Aided Ant Colony System

Comparative Analysis Reveals Conserved Protein Phosphorylation Networks Implicated in Multiple Diseases | Science Signaling

Large‐scale phosphomimetic screening identifies phospho‐modulated motif‐based protein interactions | Molecular Systems Biology

Presentation of Phospho.ELM with the example of BLAST Search feature. a... | Download Scientific Diagram

Exploring protein phosphorylation by combining computational approaches and biochemical methods - ScienceDirect
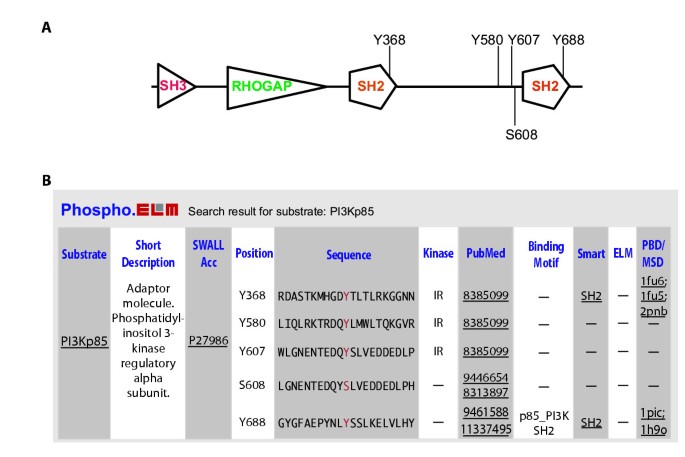
Phospho.ELM: A database of experimentally verified phosphorylation sites in eukaryotic proteins | BMC Bioinformatics | Full Text

Linear motifs and phosphorylation sites. What is a linear motif? ( in molecular biology ) - ppt download

![PhosVarDeep: deep-learning based prediction of phospho-variants using sequence information [PeerJ] PhosVarDeep: deep-learning based prediction of phospho-variants using sequence information [PeerJ]](https://dfzljdn9uc3pi.cloudfront.net/2022/12847/1/fig-4-full.png)

![PDF] qPhos: a database of protein phosphorylation dynamics in humans | Semantic Scholar PDF] qPhos: a database of protein phosphorylation dynamics in humans | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/b95803084eaa0ca510b773327e328a8212d859af/3-Figure1-1.png)



